Primer design
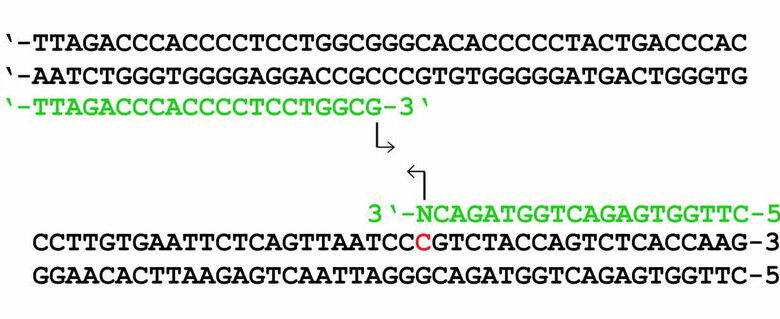
Primer design
Primers are short DNA sequences, usually composed of 18 to 24 base pair, which act as the starting point for DNA amplification by DNA polymerase enzyme during the PCR process.
Because DNA polymerase enzymes are only able to add nucleotides to the end of a DNA strand under construction, primers are essential components of the DNA replication process. Therefore, designing suitable primers is essential for performing a PCR reaction correctly and replicating the desired part with high efficiency.
To design a primer, you must first obtain the sequence of the studied gene in FASTA format from nucleic acid sequence databanks (like NCBI). In the next step, different softwares such as Oligo, gene runner, primer3, beacon designer, etc. can be used to design primers.
Basic principles of primer designing
Primer length
Typically, a length of 18 to 22 nucleotides is usually suitable for a primer. If the length of the primer is shorter, the probability of non-specific binding of the primer to the template DNA strand increases, and if the length of the primer is longer than this range, the binding of the primer to the template becomes more difficult at the annealing temperature.
Melting temperature of primers
The melting temperature of primers, or Tm, is a temperature at which half of the double-stranded DNA separates and becomes single-stranded DNA. This temperature is an estimate of the stability of the DNA-DNA hybrid and is very important in determining the annealing temperature. Typically, melting temperatures between 55 and 65 degrees are suitable for primers. The melting temperature is used to calculate the annealing temperature (Ta) of the primers.
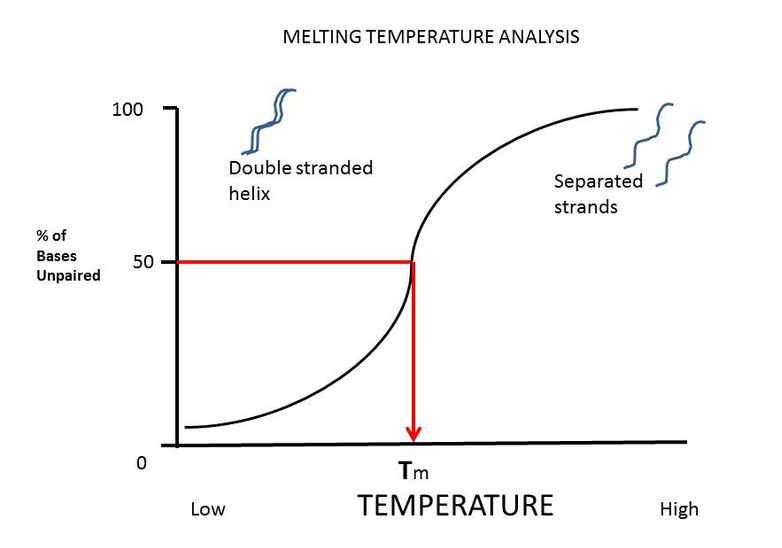
Primer connection temperature
The annealing temperature (Ta) of the primer in the PCR reaction is the temperature at which the primers bind to the template DNA. If the annealing temperature is too high, the primers and the template remain separate and no binding takes place. If the annealing temperature is too low, only some of the primer nucleotides will attach to the template and the bond does not occur properly. The annealing temperatures of both primers should be close to each other.
GC content
The ratio of GC bases to total bases in the primer sequence should be between 40-60%.
GC clamp
The presence of GC bases in the five ends of the 3 ends of the primers (called GC clamps) improves the specific connection at 3´ ends due to the stronger bond between G and C bases.
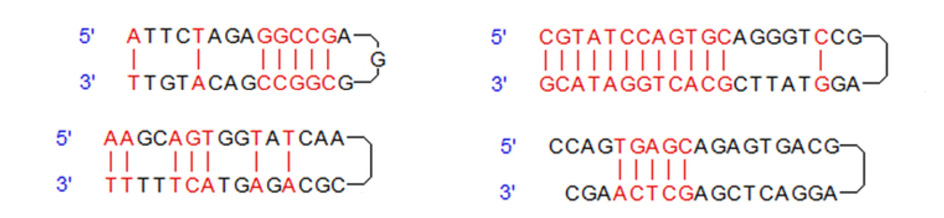
Secondary structure of primer
The formation of intramolecular secondary structures in each primer or between two primers reduces the efficiency of PCR and does not produce sufficient PCR product. These structures significantly reduce access to primers.
Hairpins
These structures are considered as the intramolecular interactions. If the 3´ end of primers has a hairpin structure with ΔG = -2 kcal /mol or a hairpin structure with ΔG = -3 kcal / mol, it is usually tolerable and does not cause a problem. (The more negative the Gibbs or ΔG free energy, the more stable hairpin structures)
Self-dimers
The self-dimer structures is caused by the interactions between the molecules of a primer. Usually, a large amount of primers is used in a reaction relative to the PCR pattern. When primers have a greater tendency to bind to themselves than a tendency to bind to the template, less PCR is produced. 3 ´ end dimers with ΔG = -5 kcal /mol and internal dimers with ΔG = -6 kcal /mol are usually tolerable.
Hetero-dimers
Heterodimers are dimers that are based on intermolecular interactions between forward and reverse primers. Typically end-to-end heterodimers with ΔG = -5 kcal /mol.
Repetition
Contains repeats of two nucleotides that are constantly repeated in a sequence. The maximum number of acceptable repeats is four repeats of two nucleotide.
Single nucleotide repeats (Runs)
Primers with high single nucleotide replications refer to a series that can cause non-specific binding. For example, AGCGGGGGGATGGGG has 5 and 4 iterations of G. The maximum number of repetitions acceptable for one base is 4 pairs of it.
Homology
Homologous regions should be avoided to increase the specificity of primers. Generally speaking, primers should be designed so that are not identified by other regions of the genome. By doing BLAST, you can ensure that the primers are specific.
Related services: RNA and DNA extraction, cDNA synthesis, primer design, PCR and real time PCR
Related content: cDNA synthesis from RNA extraction from tissue and cell – RNA Extraction from cells and tissues