Polymerase Chain Reaction PCR
Polymerase Chain Reaction PCR
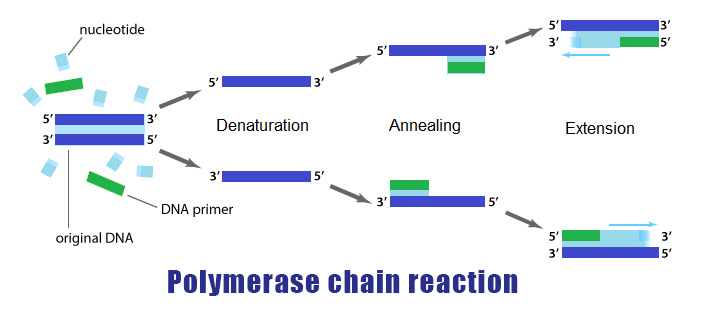
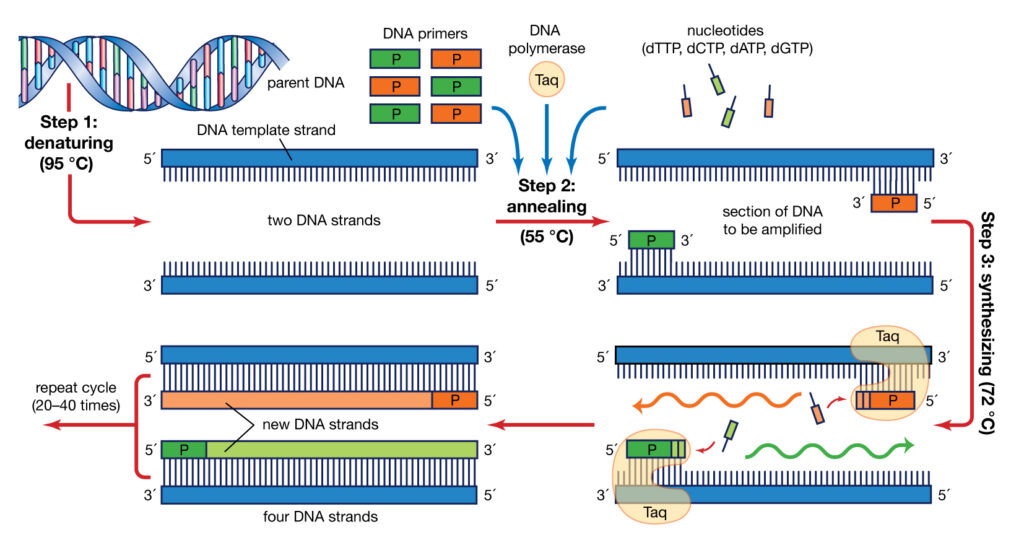
Polymerase Chain Reaction PCR is a process in which, during an interative temperature program, a specific part of the genome or cDNA (synthesized from the RNA content of a cell) is amplified by a DNA polymerase enzyme like Taq polymerase and two specific DNA sequences called primers.
The PCR reaction is scientifically very similar to the DNA replication process and was originally invented in 1983 by Kary Muliss. In this process, the DNA polymerase enzyme detects single-stranded pattern DNA from 3´ to 5´ and makes the complementary strand in 5´ to 3´.
PCR applications
- Preparation of multiple copies of the gene of interest to clone into a plasmid and express in another organism
- Assessment of presence or absence of a specific gene in a piece of DNA that is used to diagnose prenatal genetic diseases, sex determination, bacterial and viral infections, etc.
- Quantitative study of changes in gene expression under normal conditions and various diseases or treatments
expression under normal conditions and various diseases or treatments
PCR principles
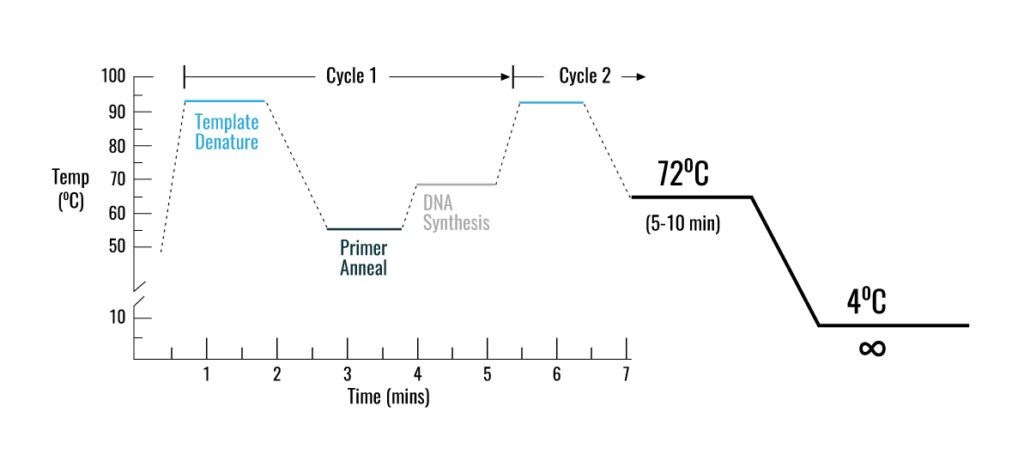
PCR method is based on a temperature program consisting of three interative steps for 30 to 40 cycles, which include the following:
1- Denaturation step
during this step, the two strands of DNA are separated by heating at 95-90 °C
2- Annealing step
at this step, the temperature reaches 53 to 65 ° C so that the primers will attach to DNA template
3- Extension step
at this step, when the temperature is suitable for the activity of DNA polymerase enzyme, the target DNA is amplified. At a temperature of about 72 °C, the heat-resistant polymerase enzyme catalyzes the polymerization reaction by consuming four types of nucleotides dATP, dGTP, dTTP, dCTP in a buffer medium containing DNA-coupled primers in the presence of MgCl2, elongating the new DNA strand. After about 30 to 40 cycles, hundreds of thousands of copies of DNA are made from very small amounts of DNA that can be identified depending on the type of PCR (traditional PCR or Real-Time PCR)
Conventional PCR
In conventional PCR or End-point PCR, after the end of the PCR reaction, PCR products can be identified as bands on the gel using agarose gel electrophoresis. This method was qualitative and the presence of the band on the gel only indicates the presence of the desired amplified gene or DNA fragment during PCR.
limitations of the traditional PCR method
- Time consuming
- Low accuracy: Viewing results is based on the size of the band observed on the gel, which may not be accurate
- Low sensitivity: Agarose gel may not be able to separate bands of small size and close to each other
- It is a qualitative method
- Post-PCR steps are numerous and include electrophoresis on agarose gel, staining the gel with ethidium bromide and observing the gel using UV device.

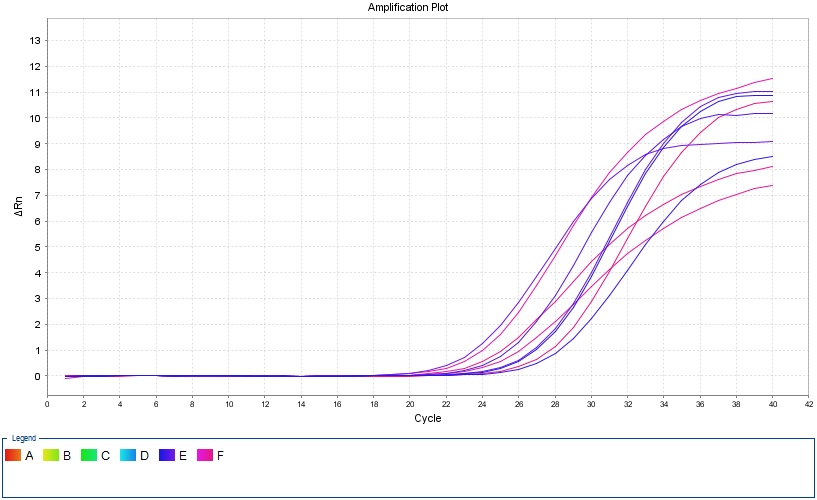
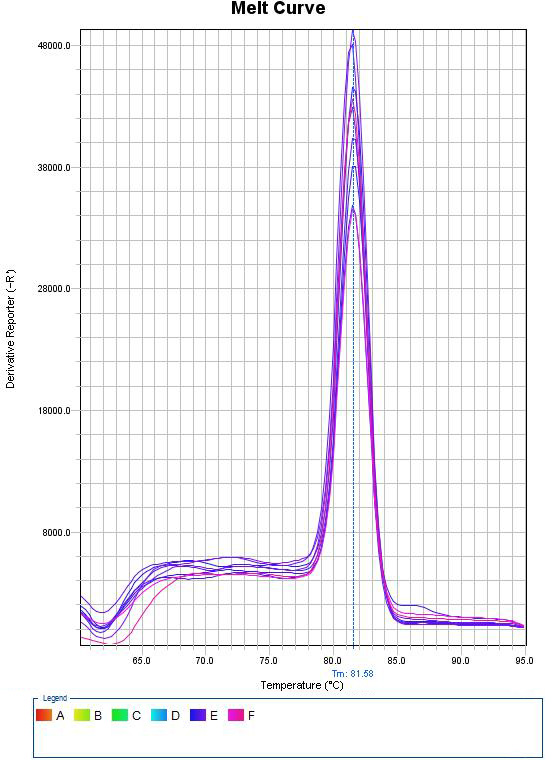
Real-Time PCR
In this method, thermocyclers equipped with fluorometric system are used, which provide the possibility of continuous monitoring of fluorescence emitted by the PCR product. This method uses fluorescent dyes that bind to double-stranded DNA or hybridization probes labeled with fluorescent dyes, which allows continuous monitoring of the PCR product without the need for agarose or polyacrylamide gel electrophoresis. This method, which was invented in 1992, was quickly accepted and widely used due to its capabilities such as reducing the time of PCR cycles, eliminating the post-PCR step and the use of fluorogenic markers and their sensitive radiation detection methods.
Real-Time PCR applications
– Determination of virus titer
– Quantitative analysis of gene expression
– Assessment of DNA damages
– Identification of pathogens
Benefits of using Real-Time PCR
– Instant observation of results during PCR process
– High sensitivity
– No post-PCR processes
Related services: RNA and DNA extraction, cDNA synthesis, primer design, PCR and real time PCR
Related content: cDNA synthesis from RNA extraction from tissue and cell – Primer design